
Imprintingdiseases
Genomic imprinting is a regulatory process that leads to monoallelic, parent-of-origin specific expression of certain genes. To date, around 100 genes have been described in humans that are subject to genomic imprinting, some of which are associated with diseases. The best known are located on chromosomes 6, 7, 11, 14, 15 and 20.
The regulation of parent-of-origin specific expression is generally linked to DNA methylation, differs in some cases between the various genes/gene clusters or is only tissue-specific.
Disorders of this regulation can be genetic or epigenetic in nature, present in cis or trans, occur in mosaic constellations or only tissue-specific and lead to various imprinting diseases depending on the affected region/affected gene. The imprinting diseases all belong to the group of rare diseases and cover a broad spectrum of clinical features and molecular genetic changes/causes (further information on diagnostics can be found in German here).
In general, we are very interested in expanding and deepening our knowledge of the regulation of expression of the different imprinting clusters and transferring this knowledge to diagnostics.
Our main focus is on
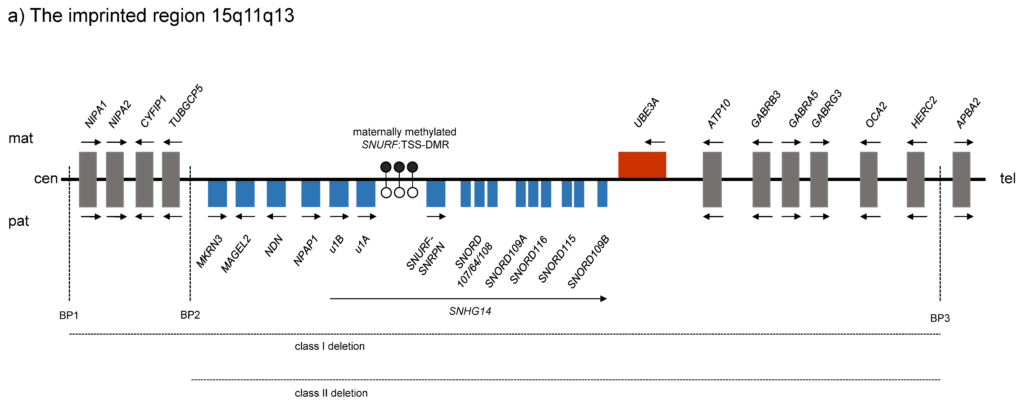
Chromosome 15q11q13
Angelman, Prader-Willi and Schaaf-Yang syndrome
Angelman and Prader-Willi syndromes are neurogenetic diseases caused by genetic and epigenetic changes on chromosome 15q11q13. The most common cause are large deletions arising from highly sequence-homologous breakpoint regions and the resulting non-allelic homologous recombination. Another cause is uniparental disomy for chromosome 15. In addition, there are a number of patients with an epigenetic change without a recognizable underlying sequence change, which are called imprinting defects (IDs).
One of our research focuses is the question of whether there are any factors that may predispose to an ID.
Another disease caused by a gene in the 15q11q13 region is Schaaf-Yang syndrome. Patients with this syndrome show a clinical phenotype that strongly overlaps with Prader-Willi syndrome. However, only truncating variants on the active paternal allele of the MAGEL2 gene lead to disease. We and others have shown that deletions of the entire gene that do not affect the imprinting centre of the region do not lead to such a phenotype.
Chromosome 14q32
Temple and Kagami-Ogata syndrome
The chromosomal region 14q32 contains an imprinting cluster that contains three DMRs and numerous maternally or paternally expressed genes. Two of the three DMRs act as imprinting centres (IC), which are essential for the regulation of parent-specific expression. We and others have shown that these two ICs act in a hierarchical manner. Not much is yet known about the function of the third DMR, which is methylated reciprocally to the other two and is hence part of our research. Various deletions have already been described by us and others as the cause of Temple or Kagami-Ogata syndrome. The detailed characterisation of such deletions is important for further research into the regulatory mechanisms within the cluster on 14q32 as well as for diagnostics
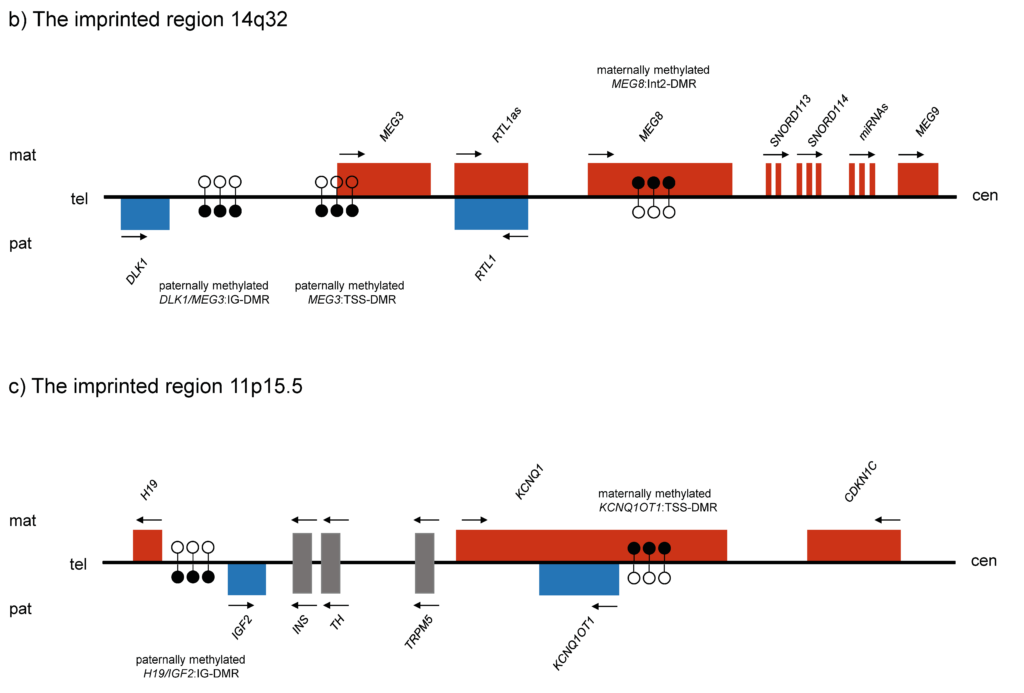
Chromosom 11p15.5
Beckwith-Wiedemann- und Silver-Russell-Syndrom
In the vast majority of cases, Beckwith-Wiedemann syndrome (BWS) and Silver-Russell syndrome (SRS) are caused by mosaic imprinting defects. In some patients, however, there is an underlying sequence change that leads to the methylation change. These include, for example, microdeletions and single nucleotide exchanges within the imprinting control region 1 (ICR1), which affect the CTCF or SOX2 binding sites. Different deletions lead to different degrees of methylation changes. Deletions affecting the KCNQ1 gene, which also lead to a methylation change but only in ICR2, must also be taken into account.
Another aspect that plays a role, particularly in patients with these syndromes, are multilocus imprinting defects, in which several imprinted loci show a methylation change. In some cases, pathogenic variants have been detected in a number of maternal effect genes in the mothers of the patients. These genes are involved in the subcortical maternal complex (SCMC). The exact mechanism of how this leads to methylation changes at multiple loci has not yet been clarified. Also, despite several disease-associated loci showing a methylation change, the BWS or SRS phenotype is usually predominant. Why this is the case is also the subject of further research.
Contact person/contact details
Dr. rer. nat.
Jasmin Beygo
Dr. rer. nat.
Deniz Kanber
Dr. rer. nat.
Sandra Ueberberg
Publications
Beygo J, Kanber D, Eggermann T, Begemann M
Molecular testing for imprinting disorders.
Medizinische Genetik 2020, 32:305-319.
Beygo J, Russo S, Tannorella P, Santen GWE, Dufke A, Schlaich E, Eggermann T
Prenatal testing for imprinting disorders: A laboratory perspective.
Prenat Diagn 2023, 43:973-982.
Marbach F, Elgizouli M, Rech M, Beygo J, Erger F, Velmans C, Stumpel C, Stegmann APA, Beck-Wodl S, Gillessen-Kaesbach G, Horsthemke B, Schaaf CP, Kuechler A
The adult phenotype of Schaaf-Yang syndrome.
Orphanet J Rare Dis 2020, 15:294.
Beygo J, Grosser C, Kaya S, Mertel C, Buiting K, Horsthemke B
Common genetic variation in the Angelman syndrome imprinting centre affects the imprinting of chromosome 15.
Eur J Hum Genet 2020, 28:835-839.
Beygo J*, Buiting K, Ramsden SC, Ellis R, Clayton-Smith J, Kanber D*
Update of the EMQN/ACGS best practice guidelines for molecular analysis of Prader-Willi and Angelman syndromes.
Eur J Hum Genet 2019, 27:1326-1340.
(*equal contribution)
Le Fevre A, Beygo J, Silveira C, Kamien B, Clayton-Smith J, Colley A, Buiting K, Dudding-Byth T
Atypical Angelman syndrome due to a mosaic imprinting defect: Case reports and review of the literature.
Am J Med Genet A 2017, 173:753-757.
Buiting K, Di Donato N, Beygo J, Bens S, von der Hagen M, Hackmann K, Horsthemke B.
Clinical phenotypes of MAGEL2 mutations and deletions.
Orphanet J Rare Dis 2014; 9:40.
Gillessen-Kaesbach G, Albrecht B, Eggermann T, Elbracht M, Mitter D, Morlot S, van Ravenswaaij-Arts CMA, Schulz S, Strobl-Wildemann G, Buiting K, Beygo J
Molecular and clinical studies in 8 patients with Temple syndrome.
Clin Genet 2018, 93:1179-1188.
Beygo J, Kuchler A, Gillessen-Kaesbach G, Albrecht B, Eckle J, Eggermann T, Gellhaus A, Kanber D, Kordass U, Ludecke HJ, Purmann S, Rossier E, van de Nes J, van der Werf IM, Wenzel M, Wieczorek D, Horsthemke B, Buiting K
New insights into the imprinted MEG8-DMR in 14q32 and clinical and molecular description of novel patients with Temple syndrome.
Eur J Hum Genet 2017.
Bens S, Kolarova J, Gillessen-Kaesbach G, Buiting K, Beygo J, Caliebe A, Ammerpohl O, Siebert R.
The differentially methylated region of MEG8 is hypermethylated in patients with Temple syndrome.
Epigenomics 2015.
Beygo J, Elbracht M, de Groot K, Begemann M, Kanber D, Platzer K, Gillessen-Kaesbach G, Vierzig A, Green A, Heller R, Buiting K, Eggermann T.
Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32.
Eur J Hum Genet 2015, 23:180-188.
Schwaibold EMC, Beygo J, Obeid K, Jauch A, Hinderhofer K, Moog U
A boy with Silver-Russell syndrome and Sotos syndrome.
Am J Med Genet A 2021, 185:549-554.
Coktu S, Spix C, Kaiser M, Beygo J, Kleinle S, Bachmann N, Kohlschmidt N, Prawitt D, Beckmann A, Klaes R, Nevinny-Stickel-Hinzpeter C, Dohnert S, Kraus C, Kadgien G, Vater I, Biskup S, Kutsche M, Kohlhase J, Eggermann T, Zenker M, Kratz CPCancer incidence and spectrum among children with genetically confirmed Beckwith-Wiedemann spectrum in Germany: a retrospective cohort study.
Br J Cancer 2020.
Beygo J, Burger J, Strom TM, Kaya S, Buiting K
Disruption of KCNQ1 prevents methylation of the ICR2 and supports the hypothesis that its transcription is necessary for imprint establishment.
Eur J Hum Genet 2019, 27:903-908.
Beygo J, Joksic I, Strom TM, Ludecke HJ, Kolarova J, Siebert R, Mikovic Z, Horsthemke B, Buiting K
A maternal deletion upstream of the imprint control region 2 in 11p15 causes loss of methylation and familial Beckwith-Wiedemann syndrome.
Eur J Hum Genet 2016, 24:1280-1286.
Beygo J, Citro V, Sparago A, De Crescenzo A, Cerrato F, Heitmann M, Rademacher K, Guala A, Enklaar T, Anichini C, Cirillo Silengo M, Graf N, Prawitt D, Cubellis MV, Horsthemke B, Buiting K, Riccio A.
The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites.
Hum Mol Genet 2013; 22:544-57.
Berland S, Appelback M, Bruland O, Beygo J, Buiting K, Mackay DJ, Karen Temple I, Houge G.
Evidence for anticipation in Beckwith-Wiedemann syndrome.
Eur J Hum Genet 2013; 21:1344-8.
Mackay DJG, Gazdagh G, Monk D, Brioude F, Giabicani E, Krzyzewska IM, Kalish JM, Maas SM, Kagami M, Beygo J, Kahre T, Tenorio-Castano J, Ambrozaityte L, Burnyte B, Cerrato F, Davies JH, Ferrero GB, Fjodorova O, Manero-Azua A, Pereda A, Russo S, Tannorella P, Temple KI, Ounap K, Riccio A, de Nanclares GP, Maher ER, Lapunzina P, Netchine I, Eggermann T, et al:
Multi-locus imprinting disturbance (MLID): interim joint statement for clinical and molecular diagnosis.
Clin Epigenetics 2024, 16:99.
Mackay D, Bliek J, Kagami M, Tenorio-Castano J, Pereda A, Brioude F, Netchine I, Papingi D, de Franco E, Lever M, Sillibourne J, Lombardi P, Gaston V, Tauber M, Diene G, Bieth E, Fernandez L, Nevado J, Tumer Z, Riccio A, Maher ER, Beygo J, Tannorella P, Russo S, de Nanclares GP, Temple IK, Ogata T, Lapunzina P, Eggermann T
First step towards a consensus strategy for multi-locus diagnostic testing of imprinting disorders.
Clin Epigenetics 2022, 14:143.
Begemann M, Rezwan FI, Beygo J, Docherty LE, Kolarova J, Schroeder C, Buiting K, Chokkalingam K, Degenhardt F, Wakeling EL, Kleinle S, Gonzalez Fassrainer D, Oehl-Jaschkowitz B, Turner CLS, Patalan M, Gizewska M, Binder G, Bich Ngoc CT, Chi Dung V, Mehta SG, Baynam G, Hamilton-Shield JP, Aljareh S, Lokulo-Sodipe O, Horton R, Siebert R, Elbracht M, Temple IK, Eggermann T, Mackay DJG
Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring.
J Med Genet 2018.
Bens S, Kolarova J, Beygo J, Buiting K, Caliebe A, Eggermann T, Gillessen-Kaesbach G, Prawitt D, Thiele-Schmitz S, Begemann M, Enklaar T, Gutwein J, Haake A, Paul U, Richter J, Soellner L, Vater I, Monk D, Horsthemke B, Ammerpohl O, Siebert R
Phenotypic spectrum and extent of DNA methylation defects associated with multilocus imprinting disturbances.
Epigenomics 2016, 8:801-816.
